Tools for quantifying separation in PCA/PLS-DA scores
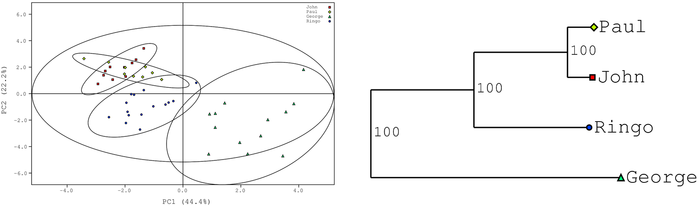
Principal Component Analysis (PCA) and Projection to Latent Structures Discriminant Analysis (PLS-DA) are two of the most widely used methods in metabolomics, chemometrics and data discovery in general. Once a validated model has been generated, separations between groups in scores space may be used to infer experimental conclusions. However, no standard method of exists to quantitatively discuss scores-space separations. We have developed a set of tools (our PCA/PLS-DA utilities, or "pca-utils" for short) that allow for quickly and quantitatively ascertaining information about group separations in scores, using both p-value calculations and bootstrap statistics to place numerical values on distances between experimental groups. Our software may be used to generate simple tables of distances or p-values, dendrograms illustrating group relationships, and scores plots annotated with 95% confidence ellipsoids informing group membership.
The PCA/PLS-DA utilities are available on GitHub at the link below. Instructions for cloning and installing from source, as well as instructions for general use, are available there.
References
Publications related to PCA/PLS-DA Utilities
- (96) B. Worley, and R. Powers* (2013) "Multivariate Analysis in Metabolomics", Current Metabolomics, 1(1):92-107 PMC44651877.
- (74) M. T. Werth, S. Halouska, M. D. Shortridge, B. Zhang and R. Powers* (2010) "Analysis of Metabolomic PCA Data using Tree Diagrams", Anal. Biochem., 399(1):58-63. PMC2824058.
Picture Gallery
From Analytical Biochemistry (2012).