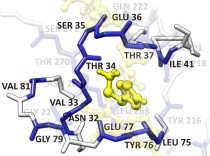
CPASSThe Comparison of Protein Active Site Structures (CPASS) database and software is used as part of our FAST-NMR assay to assign the function of a hypothetical protein or a protein of unknown function. The CPASS database and software enable the comparison of experimentally identified ligand binding sites to infer biological function and aid in drug discovery. |
PROFESSPROFESS is a biology database system that integrates databases describing PROtein Functions, Evolution, Structures and Sequences. PROFESS aims at assisting researchers in the functional and evolutionary analysis of the abundant number of novel proteins continually identified from whole-genome sequencing. |
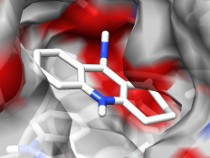
AutoDockFilterAutoDockFilter is an application to be used in concert with AutoDock which intelligently facilitates the determination of protein-ligand costructures by filtering docking results based on chemical shift perturbations from nuclear magnetic resonance experiments. |
Bioscreen DatabaseThe NMR functional chemical library is a new type of compound library that focuses on aiding in the functional annotation of novel proteins that have been identified from various ongoing genomics efforts. The NMR functional chemical library is an integral component of our FAST-NMR assay and is composed of small molecules with known biological activity. |

BORG DatabaseThe BORG database and classifier toolchain are a set of shell scripts and C utilities to perform assimilation of protein annotation data from multiple sources, and classify that set of proteins with respect to their possible relevance to a disease state, based on the assimilated annotations and a heuristic scoring function calculated from protein-protein interaction networks. |
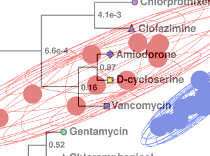
PCA/PLS-DA UtilitiesOur PCA/PLS-DA utilities allow for fast statistical quantification of group separation in scores plots resulting of PCA, PLS-DA and OPLS-DA multivariate analyses. |
MVAPACK ToolboxWe have developed an Octave/MATLAB software toolbox for processing and modeling NMR metabolomics data in an effort to centralize and streamline our 1D metabolomics methodologies around a single, customizable software platform. The code is open-source and free for any GPLv3-compliant use. |
Gap samplingWe have developed a general framework for multidimensional nonuniform sampling based on an arbitrary gap equation, as well as implemented open-source tools to generate NMR sampling schedules based on our framework. |