FAST-NMRFAST-NMR combines structural biology and NMR ligand affinity screens with bioinformatics to assign a function to a hypothetical protein or a protein of unknown function. This is based on basic tenets of biochemistry where proteins with similar functions will have similar active sites and exhibit similar ligand binding interactions, despite global differences in sequence and structure. |

MS-NMR AssayMS/NMR Assay is a protocol for rapidly screening small organic molecules for their ability to bind a target protein while obtaining structure related information as part of a structure based drug discovery and design program. The methodology takes advantage of and combines the inherent strengths of size-exclusion gel chromatography, mass spectrometry and NMR to identify bound complexes in a relatively universal high-throughput screening approach. |

Multistep NMRThe multi-step NMR assay provides structure-related information while being an integral part of a structure based drug discovery and design program. The fundamental principle of the multi-step NMR assay is to combine distinct 1D and 2D NMR techniques, in such a manner, that the inherent strengths and weakness associated with each technique is complementary to each other in the screen. |

MetabolomicsPCA is a well established statistical technique that determines the directions of largest variations in the NMR data set. The PCA data is typically presented as a two-dimensional plot where the coordinate axis corresponds to the principal components representing the directions of the two largest variations in the NMR data set. |

Optimal Mixture SizeIn order to increase the efficiency of NMR-based protein-ligand binding screens, small molecules are often screened as mixtures. An analysis of the efficiency of mixtures and a corresponding determination of the optimum mixture size that minimizes the amount of material and instrumentation time required for an NMR screen has been lacking. A model for calculating OMS based on the application of the hypergeometric distribution function to determine the probability of a “hit” for various mixture sizes and hit rates has been determined. |
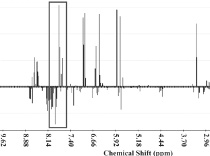
PCA NoisePrincipal component analysis (PCA) is routinely applied to the study of NMR based metabolomic data. A common concern with PCA of NMR data are the potential over emphasis of small changes in high concentration metabolites that would over-shadow significant and large changes in low-concentration components that may lead to a skewed or irrelevant clustering of the NMR data. Alleviation of this problem is obtained by simply excluding the noise region from the PCA by a judicious choice of a threshold above the spectral noise. |

Structure DeterminationThe application of deuterium labeling and residual dipolar coupling constants in combination with other structural information has demonstrated the potential for significantly expanding the range of viable protein targets for structural analysis by NMR. The analysis of NMR protein structures determined with minimal structural information is extended with a particular interest in the utility of these structures for a structure-based drug design program. |