PCA Utilities: Difference between revisions
No edit summary |
(Added to Category) |
||
| Line 131: | Line 131: | ||
[[category:Protocols]] | [[category:Protocols]] | ||
[[category:Metabolomics]] | [[category:Metabolomics]] | ||
[[category:Data_Processing_and_Analysis]] | |||
Revision as of 05:02, 11 September 2021
The PCA Utilities package provides small software routines for plotting PCA/OPLS scores and building dendrograms based on those scores. This page outlines how to install and use the pca-utils software.
Introductions
Obtaining pca-utils
You can obtain the source code to pca-utils on its GitHub page.
Installing pca-utils
The PCA utilities are a set of command line open-source UNIX/Linux programs. The software is highly portable: provided your distribution has glibc, it should compile without incident. Once you have the source code, run these commands to install it:
git clone git://github.com/geekysuavo/pca-utils.git cd pca-utils make sudo make install
By default, the programs install to /usr/bin, but you can easily change this by modifying the Makefile if you need to.
Plotting scores with ellipses
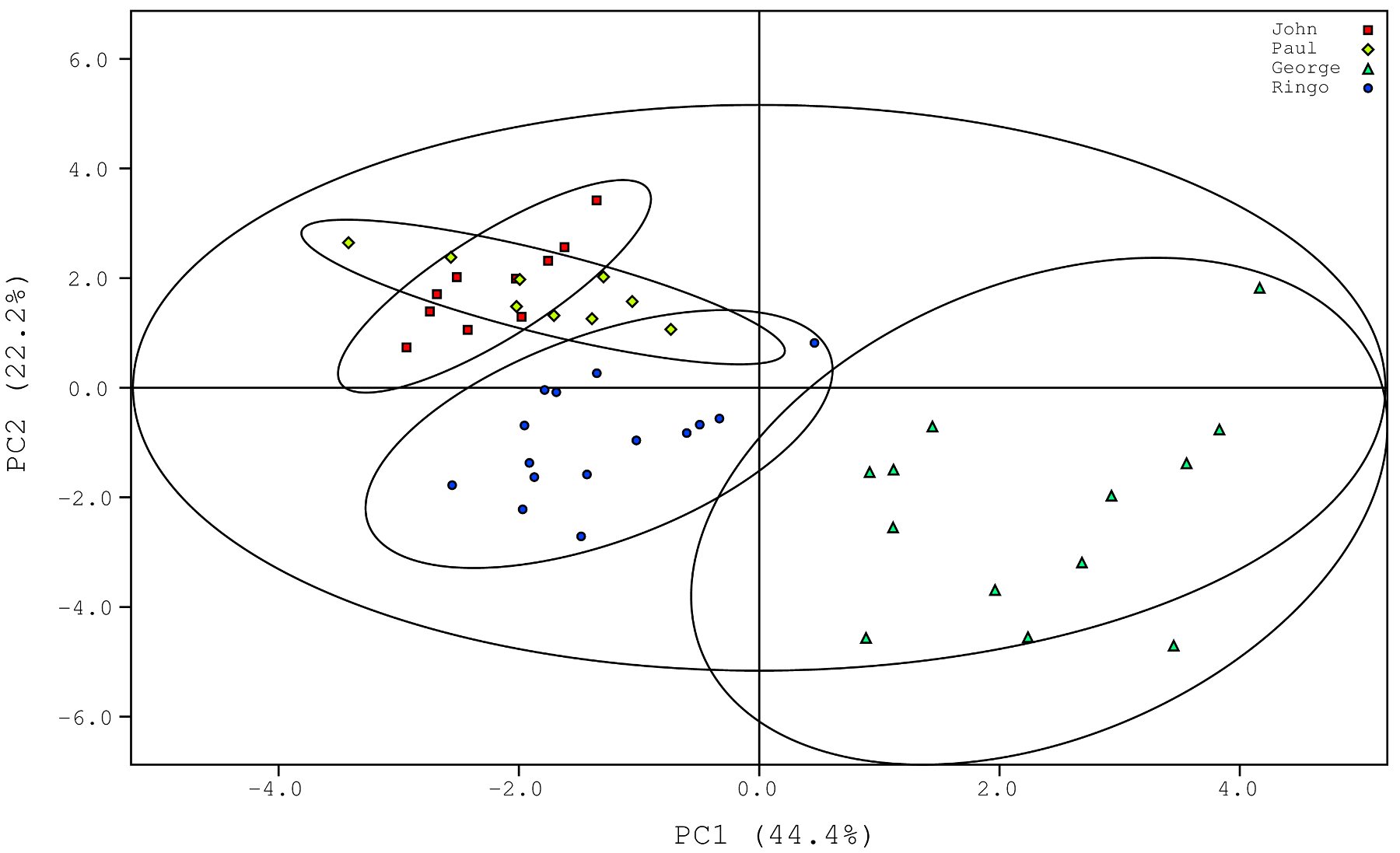
For an input list file called list.txt, you can quickly generate a postscript plot file (in this case called plot.ps) that shows your PCA scores with 95% confidence ellipses around each group:
pca-ellipses -1 44.4 -2 22.2 -i list.txt -k -o plot.ps
In the above statement, the optional arguments -1 and -2 were used to set contributions of PC1 and PC2 to 44.4% and 22.2%, respectively. You can then edit plot.ps to your liking. If you need a bit more control over your output, you can generate gnuplot-readable ellipses instead like so:
pca-ellipses -i list.txt > ellipses.txt
awk -F '\t' '/^[0-9]/{print$3,$4}' list.txt > points.txt
gnuplot> plot 'points.txt' w p, 'ellipses.txt' w l
Of course, in the second case, you're free to style everything any way you like. Happy hacking!
Plotting 3D scores with ellipsoids
That's right, ladies and gentlemen. You can plot 3D scores! Just use the pca-ellipsoids command:
pca-ellipsoids -1 44.4 -2 22.2 -3 11.1 -i list.txt > plotscript.plt
This creates a gnuplot-syntax script file that you can open up in an interactive gnuplot session like so:
gnuplot gnuplot> load 'plotscript.plt'
The above commands will open up a plot window containing the 3D plot, which you can manipulate with the mouse. Once you've found the optimal viewpoint to display the plot, copy the two view angles at the bottom left corner of the plot window. Then Add the following two lines to the top of the plotscript.plt file:
set terminal postscript enhanced color set output 'plot.ps'
Also, change view angles to your values in the following line of the plot script:
set view 60, 35
In other words, change 60 and 35 to your view angles. Finally, create the postscript file by running gnuplot again:
gnuplot plotscript.plt
You'll then have a plot.ps file that can be opened in your favorite graphics editor of choice. Happy editing!
Generating dendrograms
Two complementary methods exist for generating trees. The first uses Euclidean distances and bootstrapping statistics, while the second uses Mahalanobis distances and p-values. For datasets containing well-separated groups in scores space, the bootstrapping method will do fine. However, highly separation in overlapped data may be better quantified with p-values in many cases.
Using bootstrapping
To build a simple tree that displays to the console for quick checks, just run something like this:
pca-bootstrap -i list.txt
The default number of bootstrap iterations is 100, but pca-bootstrap can easily handle more. You can set the number of iterations to, say 1000, like so:
pca-bootstrap -i list.txt -n 1000
Everything looks good? You can save a postscript file using the -o flag:
pca-bootstrap -i list.txt -n 1000 -k -o tree.ps
Using parameterizing
To build a simple tree that displays to the console for quick checks, just run something like this:
pca-dendrogram -i list.txt
Everything looks good? You can save a postscript file using the -o flag:
pca-dendrogram -i list.txt -k -o tree.ps
If everything works, the plots generated from these methods should look something like this:
+-----------------------------------------------------------George
|3.8e-11
| +-------------------------------------Ringo
+----------------------|1.3e-08
| +-----John
+-----------------------------|0.45
+------Paul
Use of pca-bootstrap will yield values at the nodes between 0 and 100, while pca-dendrogram will yield values between 0 and 1.
Calculating p-values
If you just need p-values to accept the null hypothesis, you can use this command:
pca-overlap -i list.txt
Calculating distances
Similar to calculating p-values, raw distances may be extracted using the following command:
pca-distances -i list.txt
Other distance metrics may be supplied using the -m flag. For example, you can generate a Mahalanobis distance matrix like so:
pca-distances -m MAH -i list.txt
Calculating basic statistics
If you're interested in basic information about each group, such as mean and/or covariance, you can use pca-stats:
pca-stats -i list.txt
Goodness, that was easy, wasn't it!?
Generating random datasets
Mainly provided for entertainment value and development/debugging, pca-rand lets you generate list files that contain bivariate normally distributed point sets. Here's an example command (in the bash scripting language) to build a faux list file:
(pca-rand -H -L John -u '(-2,2)' -v '(2,0.6)' -r 45 -n 10; pca-rand -L Paul -u '(-2,2)' -v '(2,0.6)' -r 135 -n 9; pca-rand -L George -u '(3,-2)' -v '(4,3)' -r 120 -n 13; pca-rand -L Ringo -u '(-1,-1)' -v '(2,2)' -r 0 -n 15) > list.txt
No, you read that right. You can download the list file generated by a single run of this command here: File:Beatles.txt.
"Wait, I'm still confused"
Remember that every command in the pca-utils package has a help message. Just run the command that you need information on with the --help flag to get a nice message on how to use that command. You can also find manual pages for each command, e.g.:
man pca-dendrogram