Water-Suppression using pre-saturation pulses (zgpr/zgcppr): Difference between revisions
(Added to Category) |
No edit summary |
||
| Line 9: | Line 9: | ||
===ZGPR/ZGCPPR Pulse Sequence=== | ===ZGPR/ZGCPPR Pulse Sequence=== | ||
[[Image:ZGPR_ZGCPPR_pulse1.png]] | <!-- [[Image:ZGPR_ZGCPPR_pulse1.png]] --> | ||
Revision as of 21:45, 13 November 2021
Justification
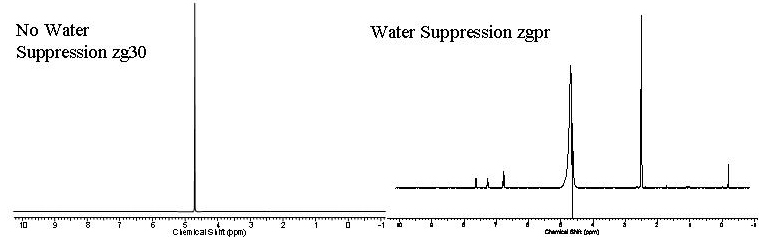
The majority of samples we run in D2O suffer from residual H2O signal masking the actual small molecule signals. This is because the small molecule concentration is typically ~100μM unlike water which has a concentration of 55M so any residual protons which have exchanged with the D2O will overpower any small molecule signal that is not of sufficient concentration. However, we can get around this issue using water suppression methods. A low power pulse at the solvent frequency and is applied during the preparation delay. This low power pulse excites the water proton signal such that no signal can fully accumulate and be measured.
Water-Suppression
The figures above displays a no water suppression zg30 spectrum compared to a water suppression zgpr spectrum respectively.
ZGPR/ZGCPPR Pulse Sequence
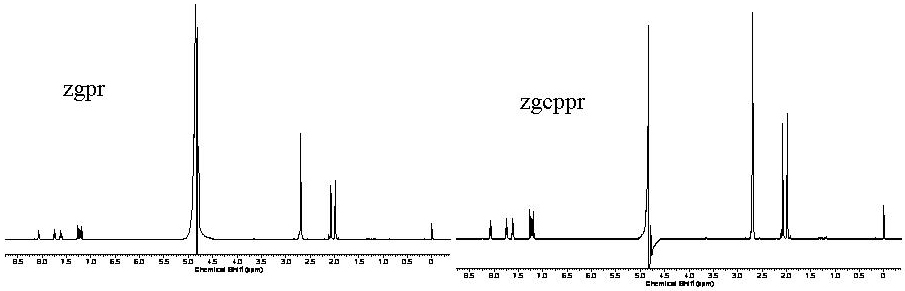
Both methods are comparable in data quality, experiment time and will suppress residual water signals. The benefit of the zgcppr pulse sequence is a more complete saturation by compensating for inhomogeneities in the applied B1 field using a series of composite 90o pulses. The result is a slightly more narrow residual water signal that must be removed during processing (zgpr: 265Hz zgcppr: 221Hz) and would be beneficial for molecules with resonances near that of water.
Methodology
Finding the O1
NOTE: The commands are for Bruker software primarily TOPSPIN.
NOTE: After any command is typed you must press enter to execute.
- We must first check where the residual water peak resonance appears in the spectrum so that we know where to apply the low power pulse to suppress the signal, this is called the O1. More generally the O1 is the center of the spectrum and is used as a reference.
- Follow steps to setup (create file, lock, shim, tune etc…) a typical 1D experiment
- Once the experiment is set up type 1H in the command line and press enter (NOTE: This will pull in the standard parameters for a typical 1D spectrum (no suppression) using the zg30 pulse sequence)
- Set number of scans (ns) to 1: Type ns and set to 1
- Set number of dummy scans (ds) to 0: Type ds and set to 0
- Set the receiver gain: Type rga (NOTE: rg should be about 10-35 check by typing rg)
- Run spectrum: Type zg
- To view FID type a
- When finished acquiring data Type efp to process
- Auto phase correct: Type apk
- Auto baseline correct: Type abs
- Move cursor over center of water peak (Very large peak ~4.5-5.0ppm) write down peak center in Hz
- Type iexpno to advance to the next experiment file
Water Suppression Pulse
- Under the new experiment file: Type pulprog and enter in the experiment you want to run (zgpr/zgcppr)
- Enter the solvent that you are using (typically D2O) Type solvent
- Pull in the standard pulse parameters (i.e. pulse lengths and delays) Type gpro (NOTE: You must have the correct solvent set to enter correct parameters)
- Type O1 and enter in new O1 value in Hertz (Hz) from the above experiment
- Set receiver gain Type rga (NOTE: rg should be about 35-50 check by typing rg)
- Set initial number of scans to 1 Type ns and set to 1
- Set initial number of dummy scans to 0 Type ds and set to 0
- Set delay/pre-saturation length: Type d1 and set to 2.00seconds (WARNING: The Cryoprobe is expensive!!! We don’t want to burn out the probe by putting in too much power. When using the Cryoprobe DO NOT set d1 above 2.00seconds with a power level no greater than 50dB!!!)
- Type zg (Note: this short experiment will let you take a quick check on the quality of water suppression)
- If water suppression is acceptable (i.e. minimal water signal) set ns and ds back to desired parameters (NOTE: Typically for our runs at 100μM small molecule ns=128 and ds=16)
- Type zg
When finished running SIGN LOG BOOK.
Transfer data to workstation or onto computers in the lab with ACD software.