FastModelFree: Difference between revisions
No edit summary |
No edit summary |
||
| Line 26: | Line 26: | ||
Ubq.in File<br /> | Ubq.in File<br /> | ||
[[Image:Ubqinputexample.jpg|500px]] <br /> | [[Image:Ubqinputexample.jpg|500px]] <br /> | ||
*Editing the Ubq.in | |||
**Second Line | |||
*** The 300 represents the number of spin systems in the R2/R1 data. You will need to change this to the correct number | |||
*** The 500.13 represents the frequency of NMR you used. For example, if you use the 600 Mhz NMR, change to 600.13. | |||
*** | |||
Revision as of 06:23, 15 October 2012
Optimizing PDB Coordinates before using Fast Model Free
PDB Inertia
This program calculates the principal moments of inertia for the atoms in a standard pdb file.. By default the program writes the moments of inertia to standard output. Optionally, the program can output a new pdb file in which the molecule is translated so that its center of mass is located at the origin and rotated so that the moments of inertia are aligned with the Cartesian axes. At present, the program only reads lines starting with the 'ATOM' keyword and only recognizes the atoms H, C, N, O, P, S.
- Type the following command in the directory containing your pdb file
- pdbinertial -r infile.pdb outfile.pdb
- This will output a translated and rotated pdb file.
R2R1 Diffusion
The program (r2r1_diffusion) uses the apprach of Tjandra, et al. [J. Am. Chem. Soc. 117:12562-12566 (1995)] to determine the diffusion tensors for spherical, and axially-symmetric motional models from experimental nitrogen-15 spin relaxation data.
Creating R2R1 file
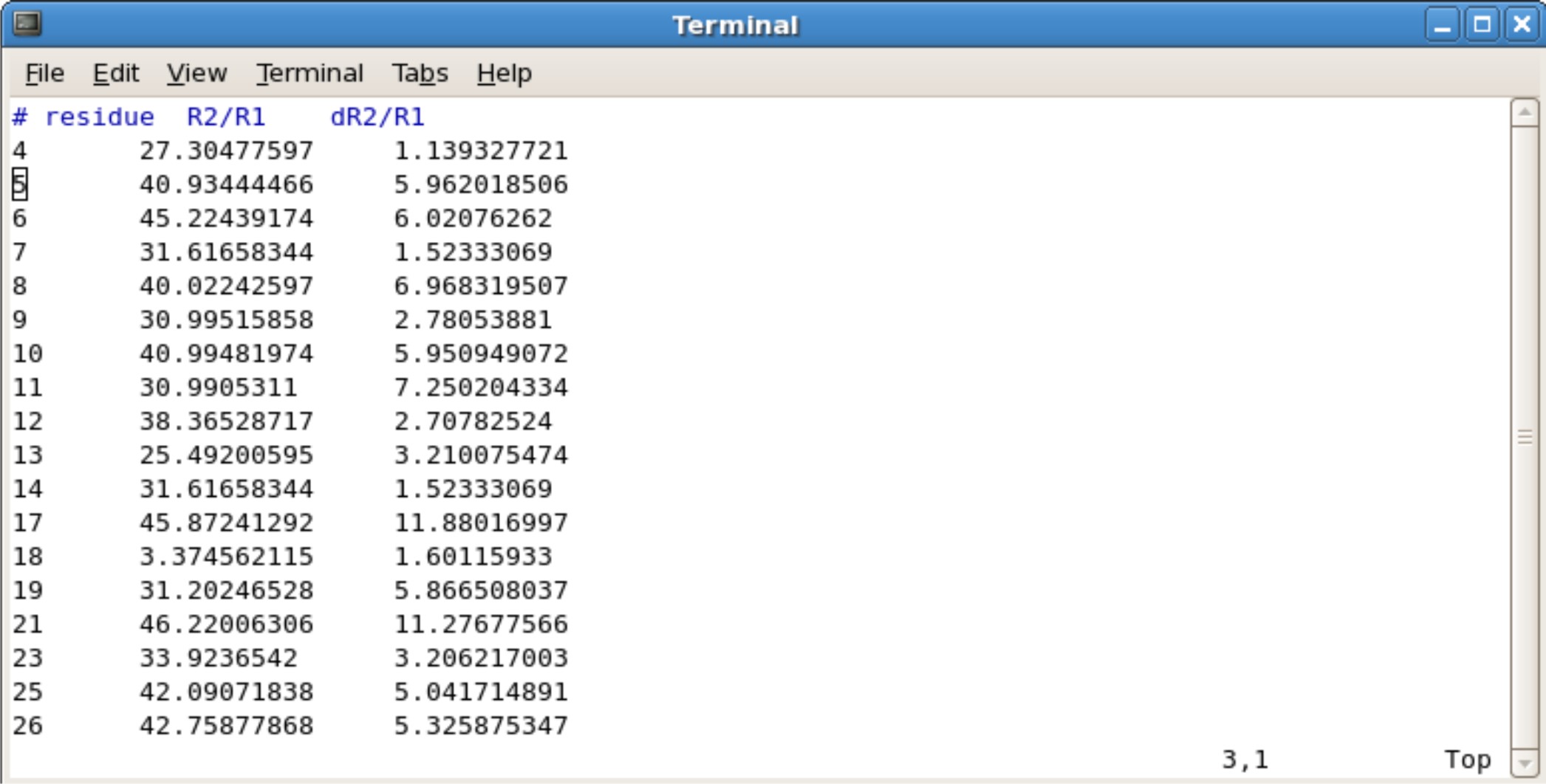
- You will need to calculate the R2/R1 ratios and R2/R1 uncertainity ratios. This can be done with any program such as excel, kaleidograph, origin, etc. You simply divide the R2 values by the R1 values for each residue. The same goes for the uncertainty values. If there is an error such as dividing by 0, remove the errors and leave blank. Save the file as a tab deliminated text file.
Setting up input file
- Copy the ubq.in file into the directory containing your translated/rotated pdb file
- The ubq.in file is located in /PROGRAMS/FMF_edited_21_02_06/r2r1_diffusion/linux
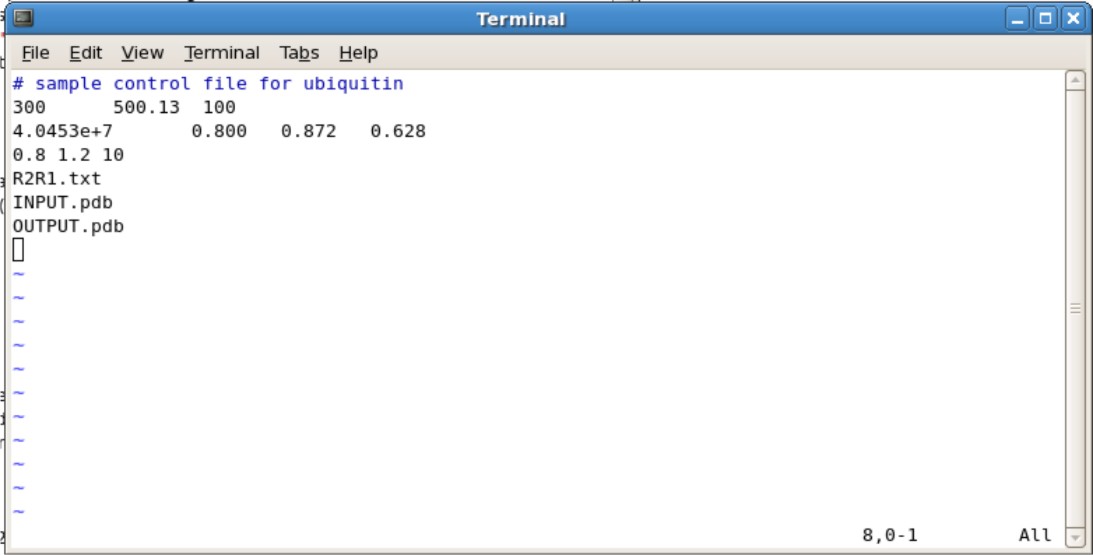
- Editing the Ubq.in
- Second Line
- The 300 represents the number of spin systems in the R2/R1 data. You will need to change this to the correct number
- The 500.13 represents the frequency of NMR you used. For example, if you use the 600 Mhz NMR, change to 600.13.
- Second Line